SONATA Simulation Configuration file¶
A simulation configuration file is a json formatted text file used for storing simulation parameters such as stimulus to drive activity, reports to collect data during run, and overrides to adapt the behavior according to experimental conditions and properties.
The file is broken up into separate sections detailed below.
version¶
Optional.
Follows the same guidelines as in Circuit Config Version and shares the same version number. The current version is “2.4”.
manifest¶
Optional.
Variables defining paths and being used in the rest of the simulation configuration file. A variable can be defined as:
“.” which is the path to the directory containing the simulation configuration file
an absolute path
“.” is always resolved as the directory containing the simulation configuration file even in the absence of manifest.
The manifest is only valid in its local file, e.g., the circuit manifest variables are not visible here.
network¶
Optional.
A parameter specifying the path of the circuit configuration file for which the simulation should be performed. The default value is “circuit_config.json”.
example:
"network": "${BASE_DIR}/circuit_config.json"
target_simulator¶
Optional.
A parameter specifying which simulator to run. Supported values : “NEURON” and “CORENEURON”. Default is “NEURON”.
node_sets_file¶
Optional.
A file defining the list of nodesets applicable to the simulation. (see SONATA Node Sets) The circuit_config nodesets are considered the base set of nodesets; the values in this file are added to the possible nodesets, and overwrite any duplicates from the base set.
node_set¶
Optional.
A parameter specifying the cells from which node set should be instantiated for the simulation. The absence of that property means that all (non virtual) nodes of all populations are loaded.
run¶
Mandatory.
Parameters defining global simulation settings. As NEURON is the engine used for simulation at BBP, find additional details about Simulation Control at https://nrn.readthedocs.io/en/latest/python/simctrl/programmatic.html
Property |
Type |
Requirement |
Description |
|---|---|---|---|
tstop |
float |
Mandatory |
Simulation runs until biological time (t) reaches tstop. Given in ms. |
dt |
float |
Mandatory |
Duration of a single integration timestep. Given in ms. |
random_seed |
integer |
Mandatory |
For random sequences, seed is a positive integer added in order to give the user the capacity to change the sequences. |
spike_threshold |
integer |
Optional |
The spike detection threshold. A spike is detected whenever the voltage in the spike detection location goes over the spike threshold value. Default is -30mV. NEURON specific details. |
integration_method |
text |
Optional |
Selects the NEURON/CoreNEURON integration method. This parameter sets the NEURON global variable h.secondorder. The allowed values are ‘0’ (default) for fully implicit backward euler, ‘1’ for Crank-Nicolson and ‘2’ for Crank-Nicolson with fixed ion currents. |
stimulus_seed |
integer |
Optional |
A non-negative integer used for seeding noise stimuli and any other future stochastic stimuli, default is 0. |
ionchannel_seed |
integer |
Optional |
A non-negative integer used for seeding stochastic ion channels, default is 0. |
minis_seed |
integer |
Optional |
A non-negative integer used for seeding the Poisson processes that drive the minis, default is 0. |
synapse_seed |
integer |
Optional |
A non-negative integer used for seeding stochastic synapses, default is 0. |
electrodes_file |
text |
Optional |
Path to the weights file describing the scaling factors for the contributions of each compartment’s transmembrane current to the LFP/EEG. Format description in sonata_tech.rst |
example:
"run": {
"tstop": 1000,
"dt": 0.025,
"random_seed": 201506,
"integration_method" : 2
}
output¶
Optional.
Parameters to override simulation output.
Property |
Type |
Requirement |
Description |
|---|---|---|---|
output_dir |
text |
Optional |
Location where output files should be written, namely spikes and reports. Relative paths are interpreted relative to location of simulation_config. Default is relative path ‘output’. |
log_file |
text |
Optional |
Specify the filename where console output is written. Default is STDOUT. (When using BBP machines, slurm will capture STDOUT file). |
spikes_file |
text |
Optional |
File name where will be listed Action Potentials generated during simulation. Default is out.h5. |
spikes_sort_order |
text |
Optional |
The sorting of the Action Potentials. Options include “none”, “by_id”, “by_time”. BBP currently only supports “none” and “by_time”. The default value is “by_time”. |
example:
"output": {
"output_dir": "output",
"spikes_file": "out.h5"
}
conditions¶
Optional.
Parameters defining global experimental conditions.
Property |
Type |
Requirement |
Description |
|---|---|---|---|
celsius |
float |
Optional |
Temperature of experiment. Default is 34.0. |
v_init |
float |
Optional |
Initial membrane voltage in mV. Default is -80. |
spike_location |
text |
Optional |
The spike detection location. Can be either ‘soma’ or ‘AIS’ for detecting spikes in either the soma or axon initial segment, respectively. Default is ‘soma’. |
extracellular_calcium |
float |
Optional |
Extracellular calcium concentration. When this parameter is provided, apply it to the synapse uHill parameter to scale the U parameter of synapses (py-neurodamus only feature). If not specified, U is set directly as read from edges file. |
randomize_gaba_rise_time |
boolean |
Optional |
When true, enable legacy behavior to randomize the GABA_A rise time in the helper functions. Default is false which will use a prescribed value for GABA_A rise time. |
mechanisms |
Optional |
Properties to assign values to variables in synapse MOD files. The format is a dictionary with keys being the SUFFIX names of MOD files (unique names of mechanisms) and values being dictionaries of variable names in the MOD files and their values. Read about NMODL2 SUFFIX description here. |
|
modifications |
Optional |
Collection of dictionaries with each member decribing a modification that mimics experimental manipulations to the circuit. |
Parameters required for modifications¶
property |
Type |
Requirement |
Description |
|---|---|---|---|
node_set |
string |
Mandatory |
Node set which receives the manipulation. |
type |
string |
Mandatory |
Name of the manipulation. Supported values are “TTX” and “ConfigureAllSections”. “TTX” mimics the application of tetrodotoxin, which blocks sodium channels and precludes spiking. “ConfigureAllSections” is a generic way to modify variables (properties, mechanisms, etc.) per morphology section. |
section_configure |
string |
Mandatory* |
For “ConfigureAllSections” manipulation, a snippet of python code to perform one or more assignments involving section attributes, for all sections that have all the referenced attributes. The wildcard %s represents each section. Multiple statements are separated by semicolons. E.g., “%s.attr = value; %s.attr2 *= value2”. |
example:
"conditions": {
"celsius": 34.0,
"spike_location": "AIS",
"mechanisms": {
"ProbAMPANMDA_EMS": {
"init_depleted": true,
"minis_single_vesicle": false
},
"ProbGABAAB_EMS" : {
"property_x": 1,
"property_y": 0.25
},
"GluSynapse": {
"property_z": "string"
}
},
"modifications": {
"applyTTX": {
"node_set": "single",
"type": "TTX"
},
"no_SK_E2": {
"node_set": "single",
"type": "ConfigureAllSections",
"section_configure": "%s.gSK_E2bar_SK_E2 = 0"
}
}
}
inputs¶
Optional.
Collection of dictionaries with each member describing one pattern of stimulus to be injected.
Property |
Type |
Requirement |
Description |
|---|---|---|---|
module |
text |
Mandatory |
The type of stimulus dictating additional parameters (see addtional tables below). Supported values: “linear”, “relative_linear”, “pulse”, “subthreshold”, “hyperpolarizing”, “synapse_replay”, “seclamp”, “noise”, “shot_noise”, “relative_shot_noise”, “absolute_shot_noise”, “ornstein_uhlenbeck”, “relative_ornstein_uhlenbeck”. |
input_type |
text |
Mandatory |
The type of the input with the reserved values : “spikes”, “extracellular_stimulation”, “current_clamp”, “voltage_clamp”, “conductance”. Should correspond according to the module (see additional tables below). Currently, not validated by BBP simulation which will use the appropriate input_type regardless of the string passed. |
delay |
float |
Mandatory |
Time in ms when input is activated. |
duration |
float |
Mandatory |
Time duration in ms for how long input is activated. |
node_set |
text |
Mandatory |
Node set which is affected by input. |
Below are additional parameters used depending on the module (input_type)
linear (current_clamp)¶
A continuous injection of current.
Property |
Type |
Requirement |
Description |
|---|---|---|---|
amp_start |
float |
Mandatory |
The amount of current initially injected when the stimulus activates. Given in nA. |
amp_end |
float |
Optional |
If given, current is interpolated such that current reaches this value when the stimulus concludes. Otherwise, current stays at amp_start. Given in nA. |
relative_linear (current_clamp)¶
A continues injection of current, regulated according to the current a cell requires to reach threshold.
Property |
Type |
Requirement |
Description |
|---|---|---|---|
percent_start |
float |
Mandatory |
The percentage of a cell’s threshold current to inject when the stimulus activates. |
percent_end |
float |
Optional |
If given, The percentage of a cell’s threshold current is interpolated such that the percentage reaches this value when the stimulus concludes. Otherwise, stays at percent_start. |
pulse (current_clamp)¶
Series of current pulse injections.
Property |
Type |
Requirement |
Description |
|---|---|---|---|
amp_start |
float |
Mandatory |
The amount of current initially injected when each pulse activates. Given in nA. |
amp_end |
float |
Optional |
If given, current is interpolated such that current reaches this value when the stimulus concludes. Otherwise, current stays at amp_start. Given in nA. |
width |
float |
Mandatory |
The length of time each pulse lasts. Given in ms. |
frequency |
float |
Mandatory |
The frequency of pulse trains. Given in Hz. |
subthreshold (current_clamp)¶
A continuous injections of current, adjusted from the current a cell requires to reach threshold.
Property |
Type |
Requirement |
Description |
|---|---|---|---|
percent_less |
integer |
Mandatory |
A percentage adjusted from 100 of a cell’s threshold current. E.g. 20 will apply 80% of the threshold current. Using a negative value will give more than 100. E.g. -20 will inject 120% of the threshold current. |
hyperpolarizing (current_clamp)¶
A hyperpolarizing current injection which brings a cell to base membrance voltage used in experiments. Note: No additional parameter are needed when using module “hyperpolarizing”. The holding current applied is defined in the cell model.
Property |
Type |
Requirement |
Description |
|---|---|---|---|
N/A |
N/A |
N/A |
N/A |
synapse_replay (spikes)¶
Spike events are created from the cells indicated in a file and delivered to their post synaptic targets. The weights of the replay synapses are set at t=0 ms and are not altered by any delayed connection.
Property |
Type |
Requirement |
Description |
|---|---|---|---|
spike_file |
text |
Mandatory |
Indicates the location of the file with the spike info for injection. |
source |
text |
Optional |
The node set to replay spikes from. |
Note: Spike files can be those output by Neurodamus. While Sonata spikes is undergoing support you may consider previous .dat files with their caveats. See .dat spike files.
seclamp (voltage_clamp)¶
Cells are held at indicated membrane voltage by injecting adapting current.
Property |
Type |
Requirement |
Description |
|---|---|---|---|
voltage |
float |
Mandatory |
Specifies the membrane voltage the targeted cells should be held at in mV. |
series_resistance |
float |
Optional |
Specifies the series resistance in M \(\Omega\). Default is 0.01 M \(\Omega\). |
noise (current_clamp)¶
Continuous injection of current with randomized deflections. Note: one must chose either “mean” or “mean_percent”.
Property |
Type |
Requirement |
Description |
|---|---|---|---|
mean |
float |
Mandatory* |
The mean value of current to inject. Given in nA. |
mean_percent |
float |
Mandatory* |
The mean value of current to inject as a percentage of a cell’s threshold current. |
variance |
float |
Optional |
The variance around the mean of current to inject using a normal distribution. |
example:
"inputs": {
"threshold_exc": {
"module": "noise",
"input_type": "current_clamp",
"mean_percent": 78,
"variance": 0.1,
"delay": 500,
"duration": 3000,
"node_set": "L5TTPC"
}
}
shot_noise, absolute_shot_noise and relative_shot_noise (current_clamp or conductance)¶
Generate a Poisson shot noise signal consisting of bi-exponential pulses with gamma distributed amplitudes occurring at exponentially distributed time intervals, resembling random synaptic input. In the Relative and Absolute versions the three parameters (rate, amp_mean, amp_var) are obtained from other three parameters: (amp_cv, mean_percent, sd_percent) for Relative and (amp_cv, mean, sigma) for Absolute, through and analytical result that connects them. In the Relative version the parameters (mean, sigma) are computed relative to a cell’s threshold current (current_clamp) or inverse input resistance (conductance), by scaling these with (mean_percent, sd_percent).
The input resistance values must be provided as an additional dataset @dynamics/input_resistance in the nodes file.
Note: fields marked Mandatory* depend on which shot_noise version is selected.
Property |
Type |
Requirement |
Description |
|---|---|---|---|
rise_time |
float |
Mandatory |
The rise time of the bi-exponential shots in ms. |
decay_time |
float |
Mandatory |
The decay time of the bi-exponential shots in ms. |
rate |
float |
Mandatory* |
For shot_noise, rate of Poisson events in Hz. |
amp_mean |
float |
Mandatory* |
For shot_noise, mean of gamma-distributed amplitudes in nA (current_clamp) or uS (conductance). |
amp_var |
float |
Mandatory* |
For shot_noise, variance of gamma-distributed amplitudes in nA^2 (current_clamp) or uS^2 (conductance). |
amp_cv |
float |
Mandatory* |
For relative_shot_noise and absolute_shot_noise, coefficient of variation (sd/mean) of gamma-distributed amplitudes. |
mean_percent |
float |
Mandatory* |
For relative_shot_noise, signal mean as percentage of a cell’s threshold current (current_clamp) or inverse input resistance (conductance). |
sd_percent |
float |
Mandatory* |
For relative_shot_noise, signal std dev as percentage of a cell’s threshold current (current_clamp) or inverse input resistance (conductance). |
mean |
float |
Mandatory* |
For absolute_shot_noise, signal mean in nA (current_clamp) or uS (conductance). |
sigma |
float |
Mandatory* |
For absolute_shot_noise, signal std dev in nA (current_clamp) or uS (conductance). |
reversal |
float |
Optional |
Reversal potential for conductance injection in mV. Default is 0. |
dt |
float |
Optional |
Timestep of generated signal in ms. Default is 0.25 ms. |
random_seed |
integer |
Optional |
Override the random seed (to introduce correlations between cells). |
ornstein_uhlenbeck and relative_ornstein_uhlenbeck (current_clamp or conductance)¶
Generate an Ornstein-Uhlenbeck process signal injected as a conductance or current. In the Relative version the parameters (mean, sigma) are computed relative to a cell’s inverse input resistance (conductance) or threshold current (current_clamp), by scaling these with (mean_percent, sd_percent).
The input resistance values must be provided as an additional dataset @dynamics/input_resistance in the nodes file.
Note: fields marked Mandatory* depend on which ornstein_uhlenbeck version is selected.
Property |
Type |
Requirement |
Description |
|---|---|---|---|
tau |
float |
Mandatory |
Relaxation time constant in ms. |
mean_percent |
float |
Mandatory* |
For relative_ornstein_uhlenbeck, signal mean as percentage of a cell’s threshold current (current_clamp) or inverse input resistance (conductance). |
sd_percent |
float |
Mandatory* |
For relative_ornstein_uhlenbeck, signal std dev as percentage of a cell’s threshold current (current_clamp) or inverse input resistance (conductance). |
mean |
float |
Mandatory* |
For ornstein_uhlenbeck, signal mean in nA (current_clamp) or uS (conductance). |
sigma |
float |
Mandatory* |
For ornstein_uhlenbeck, signal std dev in nA (current_clamp) or uS (conductance). |
reversal |
float |
Optional |
Reversal potential for conductance injection in mV. Default is 0. |
dt |
float |
Optional |
Timestep of generated signal in ms. Default is 0.25 ms. |
random_seed |
integer |
Optional |
Override the random seed (to introduce correlations between cells). |
reports¶
Optional.
Collection of dictionaries with each member describing one data collection during the simulation such as compartment voltage.
Property |
Type |
Requirement |
Description |
|---|---|---|---|
cells |
text |
Optional |
Specify which node_set to report, default is the simulation “node_set”. |
sections |
text |
Optional |
Specify which section(s) to report, available labels are dependent on the model setup. To report on all sections, use the keyword “all”. Default is “soma”. At BBP, we currently support “soma”, “axon”, “dend”, “apic”, or “all”. |
type |
text |
Mandatory |
Indicates type of data collected. “compartment”, “summation”, “synapse”, or “lfp”. Compartment means that each compartment outputs separately in the report file. Summation will sum up the values from compartments to write a single value to the report (section soma) or sum up the values and leave them in each compartment (other section types). More on summation after the table. Synapse indicates that each synapse afferent to the reported cells will have a separate entry in the report. LFP will report the contribution to the lfp (or eeg) signal from each cell, using the ‘electrodes_file’ parameter. See more after the table |
scaling |
text |
Optional |
For summation type reporting, specify the handling of density values: “none” disables all scaling, “area” (default) converts density to area values. This makes them compatible with values from point processes such as synapses. |
compartments |
text |
Optional |
For compartment type reporting, override which compartments of a section are selected to report. Options are “center” or “all”. When using “sections”:”soma”, default is “center”, for other section options, default is “all”. |
variable_name |
text |
Mandatory |
The Simulation variable to access. The variables available are model dependent. For summation type, can sum multiple variables by indicating as a comma separated strings. e.g. “ina”, “ik” |
unit |
text |
Optional |
String to output as descriptive test for unit recorded. Not validated for correctness. |
dt |
float |
Mandatory |
Interval between reporting steps in milliseconds. If assigned value smaller than simulation dt, will be set equal to simulation dt. |
start_time |
float |
Mandatory |
Time to start reporting in milliseconds. |
end_time |
float |
Mandatory |
Time to stop reporting in milliseconds. |
file_name |
text |
Optional |
Specify file name. The ‘.h5’ extension will be added if not provided. The default file name is <report_name>.h5 where ‘report_name’ is the key name of the current dictionary. |
enabled |
boolean |
Optional |
Allows for supressing a report so that it is not created. Useful for reducing output temporarily. Possible values are true/false. Default is true. |
More on type summation¶
This type of report is intended to accommodate related variables that exist in a section. For example, various electrical current sources. Depending on the sections value, the behavior of the summation adapts. Given “soma”, the values are summed across the whole cell and stored as a single value. For other sections value (e.g. “all”), values are only summed within the same compartment and stored per compartment.

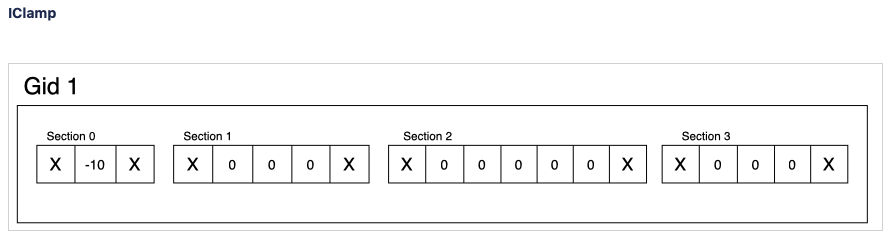
If the user has requested summation with sections soma, then the resultant single value written is [68]. Computed from (1 -10 +2+3+4+5+6+7+8+9+10+11+12).
If the user has requested summation with sections all, then the resultant data is [-9, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12 ]. Computed from (1-10, 2+0, 3+0, etc.)
example:
"reports": {
"soma": {
"cells": "Mosaic",
"sections": "soma",
"type": "compartment",
"variable_name": "v",
"unit": "mV",
"dt": 0.1,
"start_time" : 0,
"end_time" : 500,
"file_name": "soma"
"enabled" : true
},
"compartment": {
"cells": "Mosaic",
"sections": "all",
"type": "compartment",
"variable_name": "v",
"unit": "mV",
"dt": 0.1,
"start_time" : 0,
"end_time" : 500,
"file_name": "voltage"
"enabled" : true
},
"axonal_comp_centers": {
"cells": "Mosaic",
"sections": "axon",
"type": "compartment",
"variable_name": "v",
"unit": "mV",
"compartments": "center",
"dt": 0.1,
"start_time" : 0,
"end_time" : 500,
"file_name": "axon_centers"
"enabled" : true
},
"cell_imembrane": {
"cells": "Column",
"sections": "soma",
"type": "summation",
"variable_name": "i_membrane", "IClamp",
"unit": "nA",
"start_time": 0,
"end_time": 500,
"enabled": true
}
}
connection_overrides¶
Optional.
List of dictionaries to adjust the synaptic strength or other properties of edges between two sets of nodes. These are executed in the order they are read from the file. If a set of synapses are affected by multiple connection_overrides because of source and target used, the latter will overwrite any repeated fields set by a former. This is useful when making more general adjustments and then more specific adjustments. Any edges unaffected by any connection_overrides are instantiated as prescribed in the model.
Property |
Type |
Requirement |
Description |
|---|---|---|---|
name |
text |
Mandatory |
Descriptive name for the override. |
source |
text |
Mandatory |
node_set specifying presynaptic nodes. |
target |
text |
Mandatory |
node_set specifying postsynaptic nodes. |
weight |
float |
Optional |
Scalar used to adjust synaptic strength. |
spont_minis |
float |
Optional |
Synapses affected by this connection_override section will spontaneously trigger with the given rate. |
synapse_configure |
text |
Optional |
Provide a snippet of hoc code which is to be executed on the synapse objects affected by this connection_override. Use ‘%s’ to indicate where a reference to the synapse object should be filled. |
modoverride |
text |
Optional |
Changes the synapse helper files used to instantiate the synapses in this connection. A synapse helper initializes the synapse object and the parameters of the synapse model. By default, AMPANMDAHelper.hoc / GABAABHelper.hoc are used for excitatory / inhibitory synapses. The value of this field determines the prefix of the helper file to use e.g. “Newfun” would lead to NewfunHelper.hoc being used. Exceptionally, passing “GluSynapse” will lead to GluSynapse.hoc being used. That helper will use the additional parameters of the plastic synapse model read from the SONATA edges file using Neurodamus. This is required when using the GluSynapse.mod model and will fail for other models, or if the parameters are not present in the edges file. |
synapse_delay_override |
float |
Optional |
Value to override the synaptic delay time originally set in the edge file, and to be given to netcon object. Given in ms. |
delay |
float |
Optional |
Adjustments from weight of this connection_override are applied after specified delay has elapsed in ms. Note that only weight modifications are applied so all other fields (spont_minis, synapse_configure, modoverride, synapse_delay_override) are ignored. |
neuromodulation_dtc |
float |
Optional |
Only applicable to NeuroModulation projections. It overrides the |
neuromodulation_strength |
float |
Optional |
Only applicable to NeuroModulation projections. It overrides the |
example:
"connection_overrides": [
{
"name": "weaken_excitation"
"source": "Excitatory",
"target": "Mosaic,
"weight": 0.75,
"spont_minis": 0.04
},
{
"name": "deactivate_short_term_plasticity",
"source": "Mosaic",
"target": "Mosaic",
"synapse_configure": "%s.Fac = 0 %s.Dep = 0"
}
]
metadata¶
A set of variables storing remarks on the simulation, but are not used for running the simulation.
example:
"metadata": {
"note": "the first attempt at reproducing xxx experiment",
"version": "v1",
"v_int": 10,
"v_float": 0.5,
"v_bool": false
}
beta_features¶
This section is reserved for variables that are used for developing a new feature of the simulation. Once the feature goes in production, the variables should be moved to a proper section in the simulation configuration file.
example:
"beta_features": {
"v_str": "abcd",
"v_float": 0.5,
"v_int": 10,
"v_bool": false
}