SONATA Population¶
To distinguish between nodes and edges contained or connecting different brain parts, we will use populations, as per the SONATA specification. For simplicity, it is recommended to save each population in a separate file.
Naming¶
The following shows the naming adapted in BBP.
Nodes¶
For nodes, the following naming scheme is proposed:
/nodes/${part}_${type}
Where ${part} may be NCX for the neocortex, and ${type} can
presently take the following values:
neurons
astrocytes
projections
Examples:
/nodes/thalamus_astrocytes
/nodes/thalamus_neurons
/nodes/ncx_astrocytes
/nodes/ncx_neurons
/nodes/hippocampus_projections
Edges¶
Connectivity between these populations is directional, and edges will be contained in populations specifying source and target node populations as well as connection type:
/edges/${source_population}__${target_population}__${connection_type}
Note
The naming uses double underscores!
If the source and target node population are identical, a single edge population can be used. In this particular usecase the edge population is bi-directional.
Currently, the following connection types are supported:
electricalthis includes gap junctions for instance.
chemicalthis would include the following possible subtypes: glutamatergic, gabaergic, dopaminergic, serotinergic, adrenergic
synapse_astrocytethis manages the connection from an astrocyte to a particular synapse
endfootthis manages the connection between a vasculature and an astrocyte
Examples:
/edges/thalamus_neurons__ncx_neurons__chemical
Single/Multi Populations¶
In this subsection the Single-Population and Multi-Population setups are explained. We will use the following example setup with 3 nodes (A, B and C) and 4 edges (0,1,2 and 3).
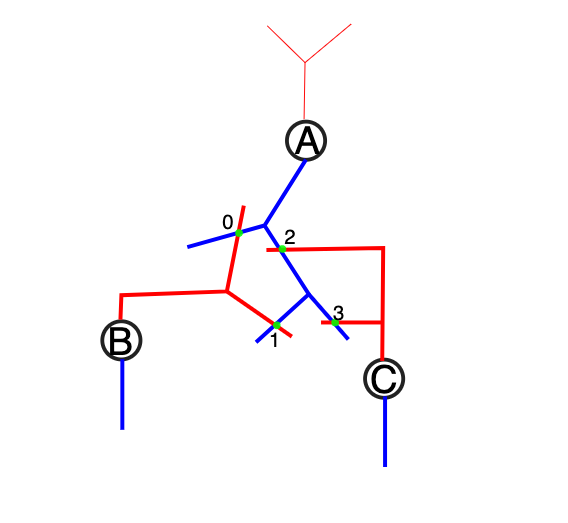
Each population in the sonata file contains a list of fields, which describe e.g the position of a node, it’s morphological type etc.
Single Population¶
The Nodes are described in a single population SingleNodePop:
/nodes/SingleNodePop/0/
node_id (index) | morphology
0 | A
1 | B
2 | C
and the edges as well in a single population SingleNodePop:
/edges/SingleEdgePop/
edge_id (index) | source_node_id | target_node_id | 0/[attributes]
| attr: 'SingleNodePop' | attr: 'SingleNodePop' |
0 | 0 | 1 | ... <- synapse 0
2 | 0 | 1 | ... <- synapse 1
3 | 0 | 2 | ... <- synapse 2
4 | 0 | 2 | ... <- synapse 3
Here, the entries source_node_id and target_node_id are lists of node_id’s,
and each of them contain an attribute attr - in this case the text SingleNodePop.
Multi Population¶
Here, the Nodes are described in different populations:
/nodes/OnlyA/0/
node_id (index) | morphology
0 | A
/nodes/OnlyB/0/
node_id (index) | morphology
0 | B
/nodes/OnlyC/0/
node_id (index) | morphology
0 | C
and so are the Edges:
/edges/A__B/
edge_id (index) | source_node_id | target_node_id | 0/[attributes]
| attr: 'OnlyA' | attr: 'OnlyB' |
0 | 0 | 0 | ... <- synapse 0
1 | 0 | 0 | ... <- synapse 1
/edges/A__C/
edge_id (index) Index | source_node_id | target_node_id | 0/[attributes]
| attr: 'OnlyA' | attr: 'OnlyC' |
0 | 0 | 0 | ... <- synapse 2
1 | 0 | 0 | ... <- synapse 3
Again, the entries source_node_id and target_node_id are lists of node_id’s.
But now, each population has different attributes attr for the entries source_node_id
and target_node_id, referring to the node population the node-id’s are taken from.
If you have \(n\) different node populations, and each population would be connected with each other population, you need \(n^2\) edge populations (as they are unidirectional, and you can have intra connections inside the node populations).
Groups¶
As per the SONATA specification it is possible to define several node groups, but within BBP we restrict to a single group 0.
Therefore, the fields node_group_id and node_group_index are not used.